Professor Jeanne Salje

Contact information
Research groups
Jeanne Salje
Associate Professor
- Group leader in bacterial cell biology
- Honorary Visiting Research Fellow in Tropical Medicine
- Wellcome Senior Research Fellow
Microbiology
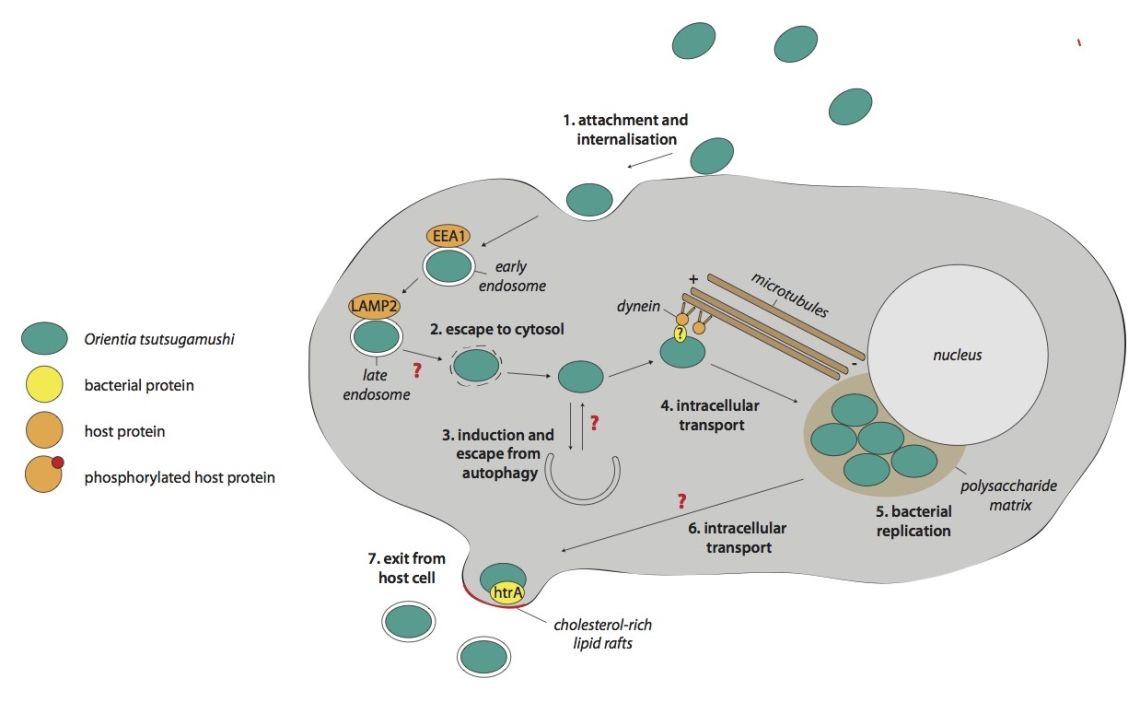
My research group studies the host-pathogen biology of the obligate intracellular bacterium Orientia tsutsugamushi. This vector-borne pathogen causes the life-threatening human disease scrub typhus, that is endemic in large parts of Asia but that is severely under-reported due to difficulties in diagnostics and surveillance. We use a combination of cell biology, biochemistry and systems biology approaches to dissect fundamental questions about the host-pathogen biology of this clinically important bacterium.
Current research is primarily focussed on the following areas:
- Developing improved experimental tools for O. tsutsugamushi research. These include methods for bacterial propagation and isolation, developing fluorescent labelling methods for light microscopy imaging and establishing methods for genetic manipulation of Orientia.
- Studying the structure of the cell wall and cell surface of O. tsutsugamushi, and understanding the implications for bacterial growth and division.
- Studying the intracellular lifecycle of O. tsutsugamushi. This includes determining molecular mechanisms of bacterial attachment and entry into host cells, escape from the endo-lysosomal pathway, and exit from infected host cells.
- Using next generation sequencing approaches to study genome organisation and dynamics. We are particularly interested in following genome changes over time and under different selective pressures.
Recent publications
-
The intracellular bacterium Orientia tsutsugamushi uses the autotransporter ScaC to activate BICD adaptors for dynein-based motility.
Journal article
Manigrasso G. et al, (2025), Nature communications, 16
-
Comparative virulence analysis of seven diverse strains of Orientia tsutsugamushi reveals a multifaceted and complex interplay of virulence factors responsible for disease.
Journal article
Chaichana P. et al, (2025), PLoS pathogens, 21
-
A comparison of super-resolution microscopy techniques for imaging tightly packed microcolonies of an obligate intracellular bacterium.
Journal article
North AJ. et al, (2024), Journal of microscopy
-
A comparison of super-resolution microscopy techniques for imaging tightly packed microcolonies of an obligate intracellular bacterium.
Preprint
North AJ. et al, (2024)