Fraydoon Rastinejad
Professor of Biochemistry and Structural Biology, Principal Investigator, Group Head and Member of Congregation
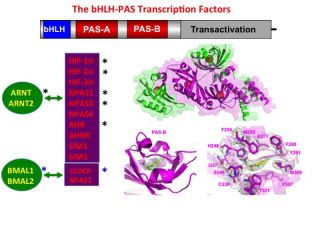
We study proteins that bind to specific DNA sequences and control the transcription of genetic information from DNA to RNA. These transcription factors, such as the human basic Helix Loop Helix-PAS (bHLH-PAS) proteins or the nuclear receptor family, can be regulated by small-molecule ligands and drug-like compounds. Members of the bHLH-PAS family heterodimerize to generate productivel transcription factors. Members include the hypoxia-inducible factors (HIF-1α, HIF-2α and HIF-3α), the aryl hydrocarbon receptor (AHR), the aryl hydrocarbon receptor repressor (AHRR), four neuronal PAS proteins (NPAS1, NPAS2, NPAS3, NPAS4), two single-minded proteins (SIM1, SIM2), and the clock circadian regulator (CLOCK).
A unifying feature of this family is their reliance on PAS domains, whose name derives from the proteins Period, ARNT, and SIM. PAS domains evolved for detecting signals in bacteria, archaea and eukaryotic organisms, and these signals can include small-molecules.
Our research program emphasises ligand discovery for this family, taking into account their individual PAS-A and PAS-B domains. We are biochemically preparing each individual PAS domain in multi-milligram amounts, and relying on innovative biochemical screening strategies that identify chemical ligands. To generate a coherent understanding of ligand-dependent actions in this family, we are attempting to link the discovery of chemical ligands to the genomic signatures of each family member. Furthermore, we are interested in addressing mechanistic questions about ligand-modulation in this family. For this purpose,we are applying structural analyses to study the stereochemical basis for ligand binding and ligand actions through induced conformational changes in these proteins.
Key publications
-
Structural integration in hypoxia-inducible factors
Journal article
Wu D. et al, (2015), Nature, 524, 303 - 308
Recent publications
-
Pharmacological targeting of BMAL1 modulates circadian and immune pathways.
Journal article
Pu H. et al, (2025), Nature chemical biology
-
Allosteric communications between domains of nuclear receptors.
Journal article
Rastinejad F., (2025), Steroids, 214
-
Decoding Allosteric Control in Hypoxia-Inducible Factors.
Journal article
Zhuang J. et al, (2023), Journal of molecular biology
-
Identification of oleoylethanolamide as an endogenous ligand for HIF-3α.
Journal article
Diao X. et al, (2022), Nature communications, 13
-
Marked and rapid effects of pharmacological HIF-2α antagonism on hypoxic ventilatory control
Journal article
Cheng X. et al, (2020), Journal of Clinical Investigation, 130, 2237 - 2251